Savu - tutorial on basic use¶
Note: The following tutorial for I13 users is out-of-date.
- How to replicate in Savu the functionality of the tomo-centre & tomo-recon commands?
- How to create a process-list file in Savu?
- How to find optimal value of CoR in Savu?
- How to reconstruct a subset of slices or the entire dataset in Savu?
- Additional remarks
- Appendices
- Appendix A: Viewing of the contents of Savu configuration files
- Appendix B: Data layout (rotation_angle, detector_y, detector_x)
- Appendix C: Viewing of the activation status of Savu processes
- Appendix D: Viewing of Savu output files produced for centring
- Appendix E: Handling of external flats and darks in Savu
- Appendix F: Viewing of Savu output files containing final-reconstruction slices
- Appendix G: Other frequently used Savu-Configurator commands
- Appendix H: Savu Tips & Tricks
This tutorial applies to Savu version 1.2 (the current default version is 2.1).
If you require Savu version 1.2, please execute: 'module add savu/1.2' (instead of 'module add savu').
How to replicate in Savu the functionality of the tomo-centre & tomo-recon commands? |
|---|
This page describes the basic use of Savu Tomography Reconstruction Pipeline for reconstructing image data in the HDF5 format. More precisely, it aims at providing some details on how to use Savu to deliver a functionality similar to that provided by the tomo-centre & the tomo-recon commands (c.f. Reconstruction from image data in the HDF format: the tomo-centre and tomo-recon commands). In particular, a brief discussion of the following typical tomography-reconstruction tasks is presented below:
- applying dark- and flat-field correction
- finding optimal centre of rotation (CoR)
- suppressing ring artefacts
- reconstructing a subset of slices or the entire dataset
For more information about Savu Pipeline and its diverse capabilities, please peruse Savu.
The conversion between the tomo-centre & the tomo-recon commands and Savu Pipeline is relatively straightforward:
| Item | tomo-centre & tomo-recon | Savu | Comment(s) |
|---|---|---|---|
| 1 | module add tomography | module add savu | Linux command for loading an appropriate module. |
| 2 | tomo-centre or tomo-recon | savu_mpi | Linux command for executing an appropriate code on the compute cluster. |
| 3 | <nexus_file> | <nexus_file> | Path to input Nexus scan file containing tomography dataset. |
| 4 | [options] | Savu process-list file | Optional arguments for executing commands in item 2. |
| 5 | <output_directory> | <output_directory> | Path to output directory. |
Similarly to the tomo-centre & tomo-recon commands, tomography reconstruction in Savu requires 3 objects: 2 input objects and 1 output object:
| Object type | Object description | Comment(s) |
|---|---|---|
| INPUT | tomography-data file | This object is provided by a path to Nexus scan file. |
| INPUT | Savu process-list (or process-configuration) file (also known as plug-in list file) | This object is provided by a path to a special Nexus file containing a list of Savu processes (also known as plug-ins) that the user intends to apply to tomography dataset in the specified order. The user can build this file from scratch or modify an existing template file to suit particular reconstruction needs. |
| OUTPUT | output directory | This object is provided by a path to a directory in which Savu can create a special Nexus output file with reconstructed slices being stored as a 3d (or possibly higher-rank) dataset. |
Savu process list is an ordered list of data-manipulation processes selected from Savu's repertoire of plug-ins to form a chain of sequential steps. Every process list in Savu requires a loader process as the initial processing step, a saver process as the final processing step, and any combination of intermediate correction, filter or reconstruction processes chained to each other and, of course, to the loader and the saver:
For example, a very basic Savu process list for standard tomography reconstruction in DLS might contain the following sequence of processes:
| Item type | Process category | Process description | Example in Savu | Comment(s) |
|---|---|---|---|---|
| INITIAL | loader | To read in raw tomography dataset from an input file. | NxtomoLoader | NxtomoLoader: this loader process reads in raw tomography dataset from standard Nexus scan files, generated in GDA. |
| INTERMEDIARY | correction(s) | To improve image quality of raw projection images with sample. | DarkFlatFieldCorrection | DarkFlatFieldCorrection: this correction process applies the classic adjustment described by equation: (projection - dark)/(flat - dark). |
| INTERMEDIARY | filter(s) | To suppress any tomography reconstruction artefacts (which would otherwise adversely affect reconstructed slices). | RavenFilter | RavenFilter (or RingRemovalWaveletfft): this filter process suppresses ring artefacts. Note that it operates on sinograms. For more details, please read Raven's paper. |
| INTERMEDIARY | reconstruction(s) | To apply a desired reconstruction algorithm, or a selection of them, to the corrected-and-filtered dataset. | AstraReconGpu | AstraReconGpu: this reconstruction process makes it possible to apply reconstruction algorithms provided by The Astra Toolbox. |
| FINAL | saver | To save final reconstructed images to an output file (or files). | Hdf5TomoSaver | Hdf5TomoSaver: this saver process saves reconstructed slices as a 3d (or possibly a higher-rank) dataset in Nexus output file. |
How to create a process-list file in Savu? |
|---|
First, a process list itself needs to be built in Savu, and then it can be saved to a Nexus file. Savu facilitates this task by providing a dedicated tool called Savu Configurator. Prior to launching Savu Configurator, the savu module needs to be made available in Linux terminal:
Then, the savu_config command can be executed to launch Savu Configurator:
The >>> prompt indicates that the above Linux session is in the Savu Configurator mode. If desired, use the help command to view a brief description of Savu Configurator's commands:
To build a basic process list for tomography reconstruction in Savu, follow the steps described below:
Add Process 1: LOADER |
|---|
Use the add command to include NxtomoLoader in the currently active, empty Savu process list as the initial, 1st process in the list:
>>> add NxtomoLoader
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : []
7) ignore_flats : None
>>>
For future reference, note that the output from the add (and many other reporting-back) command(s) has the following general form:
<process index>) <process name>
<process-parameter index>) <parameter name> : <parameter value>
...
<process-parameter index>) <parameter name> : <parameter value>
...
[<process index>) <process name>
<process-parameter index>) <parameter name> : <parameter value>
...
<process-parameter index>) <parameter name> : <parameter value>]
The above <process index> and <process-parameter index> are used for referring to, and manipulating, processes in the currently active process list in Savu Configurator.
Add Process 2: DARK-FLAT-FIELD CORRECTION |
|---|
Use the add command to include DarkFlatFieldCorrection in the currently active Savu process list as the next (i.e. 2nd) process in the list:
>>> add DarkFlatFieldCorrection
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : []
7) ignore_flats : None
2) DarkFlatFieldCorrection
1) in_datasets : []
2) out_datasets : []
3) pattern : PROJECTION
>>>
Add Process 3: RAVEN FILTER |
|---|
Use the add command to include RavenFilter in the currently active Savu process list as the next (i.e. 3rd) process:
>>> add RavenFilter
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : []
7) ignore_flats : None
2) DarkFlatFieldCorrection
1) in_datasets : []
2) out_datasets : []
3) pattern : PROJECTION
3) RavenFilter
1) in_datasets : []
2) nvalue : 4
3) out_datasets : []
4) vvalue : 2
5) padFT : 20
6) uvalue : 20
>>>
Add Process 4: ASTRA RECON GPU |
|---|
Use the add command to include AstraReconGpu in the currently active Savu process list as the next (i.e. 4th) process in the list:
>>> add AstraReconGpu
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : []
7) ignore_flats : None
2) DarkFlatFieldCorrection
1) in_datasets : []
2) out_datasets : []
3) pattern : PROJECTION
3) RavenFilter
1) in_datasets : []
2) nvalue : 4
3) out_datasets : []
4) vvalue : 2
5) padFT : 20
6) uvalue : 20
4) AstraReconGpu
1) in_datasets : []
2) init_vol : None
3) FBP_filter : ram-lak
4) res_norm : False
5) center_of_rotation : 0.0
6) log : True
7) out_datasets : []
8) number_of_iterations : 1
9) preview : []
10) reconstruction_type : FBP_CUDA
11) sino_pad : True
>>>
Add Process 5: SAVER |
|---|
Use the add command to include Hdf5TomoSaver in the currently active Savu process list as the next (i.e. 5th) and final process in the list:
>>> add Hdf5TomoSaver
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : []
7) ignore_flats : None
2) DarkFlatFieldCorrection
1) in_datasets : []
2) out_datasets : []
3) pattern : PROJECTION
3) RavenFilter
1) in_datasets : []
2) nvalue : 4
3) out_datasets : []
4) vvalue : 2
5) padFT : 20
6) uvalue : 20
4) AstraReconGpu
1) in_datasets : []
2) init_vol : None
3) FBP_filter : ram-lak
4) res_norm : False
5) center_of_rotation : 0.0
6) log : True
7) out_datasets : []
8) number_of_iterations : 1
9) preview : []
10) reconstruction_type : FBP_CUDA
11) sino_pad : True
5) Hdf5TomoSaver
>>>
This completes the task of building a basic Savu process list for tomography reconstruction. It is a good practice to execute the disp command to display the list's contents for final verification before saving it to a file:
Save Process Chain to file |
|---|
Now, use the save command to store the currently active Savu process list in a Nexus file in an accessible directory of your choice, here /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs (note that the destination directory must already exist, and that the save command overwrites any existing files with the same name):
Savu Configurator command: save <filename>.nxs
>>> save /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
Are you sure you want to save the current data to '/dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs' [y/N]y
Saving file /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
If desired, the exit command can now be invoked to leave Savu Configurator to return to the original (savu-enabled) Linux session
>>> exit Are you sure? [y/N]y Thanks for using the application (savu_v1.2) ~>
which in turn enables one to explicitly verify that the tomo_recon_base_config.nxs file can indeed be found in the /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/ directory:
Linux command: ls -l
(savu_v1.2) ~>ll /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
-rw-rw----+ 1 vxu94780 vxu94780 34216 Mar 1 13:44 /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
(savu_v1.2) ~>
Incidentally, note that all the processes in the above tomo_recon_base_config.nxs file are on their default settings. See Appendix A for information about viewing and interpreting the contents of Savu process-list files.
How to find optimal value of CoR in Savu? |
|---|
As in the case of the tomo-centre command, an optimal value of CoR can be determined by visual inspection of a number of trial reconstructions of a test slice (or a set of test slices). Similarly to the tomo-centre command, one needs to specify the index of a test slice (or the indices of a set of test slices) and a list of trial CoRs. These additional parameters can be specified in Savu by modifying appropriate fields in tomo_recon_base_config.nxs. In what follows, /dls/mx-scratch/tomo/2013/cm12345-1/13429_subx.nxs is used as an example Nexus scan file (c.f. Session A. DAWN Training - Tomography) for finding an optimal CoR by reconstructing slice with index 1093 with the following 5 trial values for CoR:
1919.8 (= 2019.8 - 2*50.0)
1969.8 (= 2019.8 - 50.0)
2019.8 (this particular value is known to be optimal for this tomography dataset)
2069.8 (= 2019.8 + 50.0)
2119.8 (= 2019.8 + 2*50.0)
If not already available, launch Savu Configurator (see the top of this page), and then use the open command to load the previously-created tomo_recon_base_config.nxs file into Savu Configurator:
Note that, at this particular stage, all 5 processes in the above tomo_recon_base_config.nxs file are on their default settings. Now, use the mod command to modify the preview parameter (<process-parameter index> = 6) of the NxtomoLoader process (<process index> = 1) so that a single slice, indexed 1093, is loaded for reconstruction (incidentally, the default setting of NxtomoLoader's preview parameter (i.e, [ ]) is to load in all slices). Since the shape of the tomography dataset in /dls/mx-scratch/tomo/2013/cm12345-1/13429_subx.nxs has the form (<tomography-rotation-angle-enumeration>, <raw-image-y-axis>, <raw-image-x-axis>) (see Appendix B for more details), one needs to select the middle index of the preview parameter to be 1093 and specify the other two indices so as to exhaust their respective full ranges of values.
Incidentally, the general syntax for selecting slicing parameters for preview has the form:
<start>:<stop>:<step>:<chunk>
where each of the four components should be replaced with an integer or the key words ‘end’ or ‘mid’ (note that the <stop> slice does not get included in the resulting subset). In the case at hand, this leads to the following mod-ing:
>>> mod 1.6 [:, 1093, :]
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : [:, 1093, :]
7) ignore_flats : None
>>>
Note that expression [0:end:1:1, 1093:1093+1:1:1, 0:end:1:1] is equivalent in Savu Configurator to a more-compact expression [0:end:1:1, 1093, 0:end:1:1] and to the most-compact [:, 1093, :].
As for the desired 5 centring values, 1919.8, 1969.8, 2019.8, 2069.8, 2119.8, they can be selected via the center_of_rotation parameter (<process-parameter index> = 5) of the AstraReconGpu process (<process index> = 4) in a one-by-one fashion:
>>> mod 4.5 1919.8;1969.8;2019.8;2069.8;2119.8
4) AstraReconGpu
1) in_datasets : []
2) init_vol : None
3) FBP_filter : ram-lak
4) res_norm : False
5) center_of_rotation : 1919.8;1969.8;2019.8;2069.8;2119.8
6) log : True
7) out_datasets : []
8) number_of_iterations : 1
9) preview : []
10) reconstruction_type : FBP_CUDA
11) sino_pad : True
>>>
or as an arithmetic series, specified with syntax:
<first-value>:<last-value>:<difference>;
Please note the trailing semicolon! Also note that <last-value> is included in the resulting numerical sequence:
>>> mod 4.5 2019.8-2*50.0:2019.8+2*50.0:50.0;
4) AstraReconGpu
1) in_datasets : []
2) init_vol : None
3) FBP_filter : ram-lak
4) res_norm : False
5) center_of_rotation : 2019.8-2*50.0:2019.8+2*50.0:50.0;
6) log : True
7) out_datasets : []
8) number_of_iterations : 1
9) preview : []
10) reconstruction_type : FBP_CUDA
11) sino_pad : True
>>>
To speed up the task of finding an optimal CoR, it is advantageous to deactivate the RavenFilter process (<process index> = 3) by executing the mod command with the following self-explanatory syntax:
<process index>.<off | on>
In the case at hand, this gives:
>>> mod 3.off
switching plugin 3 OFF
***OFF*** 3) RavenFilter
1) in_datasets : []
2) nvalue : 4
3) out_datasets : []
4) vvalue : 2
5) padFT : 20
6) uvalue : 20
>>>
See Appendix C for information on how to view the results of this modification in hdfview.
Finally, the above modifications of /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs can conveniently be saved to a new file, named /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config_raven_off.nxs
Savu Configurator command: save <path-to-process-list-file>
>>> save /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config_raven_off.nxs
Are you sure you want to save the current data to '/dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config_raven_off.nxs' [y/N]y
Saving file /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config_raven_off.nxs
>>>
Now, exit Savu Configurator to return to the original (savu-enabled) Linux session:
>>> exit Are you sure? [y/N]y Thanks for using the application (savu_v1.2) ~>
The savu_mpi command for CoR optimisation |
|---|
The trial reconstructions are carried out by executing the savu_mpi command, using the following 3-argument syntax (note that the output directory is automatically created if it doesn't already exist):
savu_mpi <path-to-Nexus-scan-file> <path-to-Savu-process-list-file> <path-to-output-directory>
In the case at hand, this leads to the following:
Savu command: savu_mpi
(savu_v1.2) ~>savu_mpi /dls/mx-scratch/tomo/2013/cm12345-1/13429_subx.nxs /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config_raven_off.nxs /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/
Loading 64-bit Anaconda Python, version 4.2.13
The Savu path is: /dls_sw/apps/savu/anaconda/envs/savu_v1.2/lib/python2.7/site-packages/savu-1.2-py2.7.egg
Creating the output folder /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301145340_13429_subx
************************************************************************
*** THANK YOU FOR USING SAVU! ***
Your job has been submitted to the cluster with job number 17902742.
* Monitor the status of your job on the cluster:
>> module load global/cluster
>> qstat
* Monitor the progression of your Savu job:
>> tail -f /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301145340_13429_subx/user.log
>> Ctrl+C (to quit)
For a more detailed log file see:
/dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301145340_13429_subx/savu.o17902742
************************************************************************
(savu_v1.2) ~>
As the above output from the savu_mpi command suggests, one should wait until the relevant cluster jobs are finished before examining any output images. The actual output directory is a time-stamped sub-directory of the command-line, nominal output directory, /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/. In this particular case, all the output files can be found inside the 20170301145340_13429_subx sub-directory, with the directory name being generated from template <YYYY><MM><DD><hh><mm><ss>_<Nexus-scan-filename>. The actual output directory contains a number of different files whose names are self-explanatory:
(savu_v1.2) ~>ll /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/ total 0 drwxrwx---+ 2 vxu94780 vxu94780 512 Mar 1 14:56 20170301145340_13429_subx (savu_v1.2) ~> (savu_v1.2) ~> (savu_v1.2) ~>ll /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/20170301145340_13429_subx/ total 346496 -rw-r-----+ 1 vxu94780 vxu94780 737143 Mar 1 14:58 savu.o17902742 -rw-r-----+ 1 vxu94780 vxu94780 0 Mar 1 14:53 savu.po17902742 -rw-rw----+ 1 vxu94780 vxu94780 41752 Mar 1 14:58 subx_processed.nxs -rw-rw----+ 1 vxu94780 vxu94780 29376432 Mar 1 14:58 tomo_p1_dark_flat_field_correction.h5 -rw-rw----+ 1 vxu94780 vxu94780 324153088 Mar 1 14:58 tomo_p2_astra_recon_gpu.h5 -rw-rw----+ 1 vxu94780 vxu94780 1594 Mar 1 14:58 user.log (savu_v1.2) ~>
The subx_processed.nxs file is the principal output file for this Savu reconstruction, the filename being derived from template <Nexus-scan-filename>_processed.nxs. The 5 trial reconstructions are stored in the /entry/final_result_tomo dataset. Note that this subx_processed.nxs contains links to 2 external HDF5 files:
tomo_p1_dark_flat_field_correction.h5 (dark-and-flat-field-corrected dataset)
and
tomo_p2_astra_recon_gpu.h5 (reconstructed images).
Note also that /entry/final_result_tomo is in fact a link to the 2-AstraReconGpu-tomo group inside the tomo_p2_astra_recon_gpu.h5 file. These links can easily be identified in the following output from the h5dump -n command (do not forget to include the n-option or otherwise all datasets will be printed to the screen as long arrays of numbers!):
See Appendix D for screenshots of the contents of a selection of centring output files, viewed in hdfview and DAWN.
How to reconstruct a subset of slices or the entire dataset in Savu? |
|---|
The functionality provided by the tomo-recon command can easily be replaced by that furnished by the savu_mpi command. If not already available, launch Savu Configurator (see the top of this page), and then use the open command to load the previously-created, all-default-settings tomo_recon_base_config.nxs file into Savu Configurator:
To reconstruct a (contiguous) slab of, say, 4 consecutive slices from (and including) slice with index 1093 to (and including) slice with index 1096 (=1093+4-1), apply the mod command to the preview parameter (<process-parameter index> = 6) of the NxtomoLoader process (<process index> = 1) in the following way:
>>> mod 1.6 [0:end:1:1, 1093:1096+1:1:1, 0:end:1:1]
1) NxtomoLoader
1) flat : [None, None, 1]
2) 3d_to_4d : False
3) data_path : entry1/tomo_entry/data/data
4) dark : [None, None, 1]
5) angles : None
6) preview : [0:end:1:1,1093:1096+1:1:1,0:end:1:1]
7) ignore_flats : None
>>>
Incidentally, to reconstruct the entire dataset, leave the preview parameter of NxtomoLoader in its default setting, i.e. [ ].
Then, select CoR to be 2019.8 (the optimal value identified earlier during the centring task) by mod-ing the center_of_rotation parameter (<process-parameter index> = 5) of the AstraReconGpu process (<process index> = 4):
>>> mod 4.5 2019.8
4) AstraReconGpu
1) in_datasets : []
2) init_vol : None
3) FBP_filter : ram-lak
4) res_norm : False
5) center_of_rotation : 2019.8
6) log : True
7) out_datasets : []
8) number_of_iterations : 1
9) preview : []
10) reconstruction_type : FBP_CUDA
11) sino_pad : True
>>>
It is a good practice to use the disp command to verify the process list before saving it to a file:
Note that the RavenFilter process (<process-index> = 3) is now ON. If desired, use the disp <process-index> -v command (the v-option stands for verbose) to obtain more information about any relevant process parameters:
>>> disp 3 -v
3) RavenFilter(savu.plugins.filters.raven_filter)
A plugin to remove ring artefacts
1) in_datasets : []
A list of the dataset(s) to process.
2) nvalue : 4
To define the shape of filter.
3) out_datasets : []
A list of the dataset(s) to create.
4) vvalue : 2
How many rows to be applied the filter.
5) padFT : 20
Padding for Fourier transform.
6) uvalue : 20
To define the shape of filter, e.g. bad=10, moderate=20, minor=50.
>>>
In the case of the RavenFilter process, the default value of 20 for the uvalue parameter is recommended to suppress ring artefacts of moderate severity, and this particular value appears reasonable to apply in the case of /dls/mx-scratch/tomo/2013/cm12345-1/13429_subx.nxs. However, note that the uvalue parameter can be specified to be a set of values, e.g. a triple of numbers 15;20;23. Similarly, note that the default value of the reconstruction_type parameter (<process-parameter index> = 10) of the AstraReconGpu process (<process index> = 4) is FBP_CUDA, but this parameter can also be specified to be a set of values, e.g. FBP_CUDA;CGLS_CUDA. As before, use the disp <process-index> -v command to obtain more information about all options available for AstraReconGpu:
Now, use the save command to store all the above modifications of /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs in the same file:
Savu Configurator command: save <path-to-process-list-file>
>>> save /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
Are you sure you want to save the current data to '/dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs' [y/N]y
Saving file /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs
>>>
Then, exit Savu Configurator to return to to the original (savu-enabled) Linux session:
>>> exit Are you sure? [y/N]y Thanks for using the application (savu_v1.2) ~>
The savu_mpi command for running full (or partial) reconstruction |
|---|
Finally, the selected slab of 4 slices can be reconstructed, using the previously-determined optimal CoR value of 2019.8, with the help of the following savu_mpi command:
Savu command: savu_mpi <path-to-Nexus-scan-file> <path-to-Savu-process-list-file> <path-to-output-directory>
(savu_v1.2) ~>savu_mpi /dls/mx-scratch/tomo/2013/cm12345-1/13429_subx.nxs /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/
Loading 64-bit Anaconda Python, version 4.2.13
The Savu path is: /dls_sw/apps/savu/anaconda/envs/savu_v1.2/lib/python2.7/site-packages/savu-1.2-py2.7.egg
Creating the output folder /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301171639_13429_subx
************************************************************************
*** THANK YOU FOR USING SAVU! ***
Your job has been submitted to the cluster with job number 17905569.
* Monitor the status of your job on the cluster:
>> module load global/cluster
>> qstat
* Monitor the progression of your Savu job:
>> tail -f /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301171639_13429_subx/user.log
>> Ctrl+C (to quit)
For a more detailed log file see:
/dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301171639_13429_subx/savu.o17905569
************************************************************************
(savu_v1.2) ~>
As the above output from the savu_mpi command suggests, one should wait until the relevant cluster jobs are finished before examining any output images. As in the case of the centring task, the actual output directory is a time-stamped sub-directory, called 20170301171639_13429_subx, of the command-line, nominal output directory, /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/; it contains the following files:
(savu_v1.2) ~>ll /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429/ total 0 drwxrwx---+ 2 vxu94780 vxu94780 512 Mar 1 14:56 20170301145340_13429_subx drwxrwx---+ 2 vxu94780 vxu94780 512 Mar 1 17:19 20170301171639_13429_subx (savu_v1.2) ~> (savu_v1.2) ~> (savu_v1.2) ~> (savu_v1.2) ~>ll /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/13429//20170301171639_13429_subx total 486016 -rw-r-----+ 1 vxu94780 vxu94780 1021308 Mar 1 17:19 savu.o17905569 -rw-r-----+ 1 vxu94780 vxu94780 0 Mar 1 17:16 savu.po17905569 -rw-rw----+ 1 vxu94780 vxu94780 46624 Mar 1 17:19 subx_processed.nxs -rw-rw----+ 1 vxu94780 vxu94780 116186248 Mar 1 17:19 tomo_p1_dark_flat_field_correction.h5 -rw-rw----+ 1 vxu94780 vxu94780 116414536 Mar 1 17:19 tomo_p2_raven_filter.h5 -rw-rw----+ 1 vxu94780 vxu94780 259425984 Mar 1 17:19 tomo_p3_astra_recon_gpu.h5 -rw-rw----+ 1 vxu94780 vxu94780 1415 Mar 1 17:19 user.log (savu_v1.2) ~>
As before, the subx_processed.nxs file is the principal Nexus output file, with the set of 4 reconstructed slices being stored in the /entry/final_result_tomo dataset. Note that this subx_processed.nxs contains links to 3 external HDF5 files:
tomo_p1_dark_flat_field_correction.h5 (dark-and-flat-field-corrected dataset),
tomo_p2_raven_filter.h5 (ring-artefact suppressed dataset),
and
tomo_p3_astra_recon_gpu.h5 (reconstructed images).
Note also that /entry/final_result_tomo is in fact a link to the 3-AstraReconGpu-tomo group inside tomo_p3_astra_recon_gpu.h5. As before, these links can easily be identified using the h5dump -n command (do not forget to include the n-option or otherwise all datasets will be printed to the screen as long arrays of numbers!):
See Appendix F for screenshots of the contents of a selection of final-reconstruction output files, viewed in hdfview and DAWN.
Additional remarks |
|---|
Tips for more advanced use of Savu |
|---|
Note that Savu contains a dedicated plug-in, called VoCentering, for finding an optimal value of CoR automatically. This filter process would typically be added to a tomography process list just before the reconstruction process, e.g. AstraReconGpu. Another filter process worth mentioning is PaganinFilter (this process would typically be included after any ring-artefacts-suppression process but before VoCentering).
An additional selection of frequently used and handy Savu-Configurator commands can be found in Appendix G, whereas Appendix H contains a selection of Savu Tips & Tricks.
Extraction of TIFF images and their bit-depth reduction |
|---|
For more information, please see Extraction of TIFF images from image data in the HDF format (with optional bit-depth reduction) and related matters.
Appendices |
|---|
Appendix A: Viewing of the contents of Savu configuration files |
|---|
The contents of any Savu process-list file can be viewed in the HDFView application or in DAWN (using either the File Browsing or DExplore perspective). However, if you opt for DAWN (recommended) and intend to use it in the same Linux terminal as Savu, then it is essential to execute 'module add savu' after 'module add dawn' (otherwise Savu might not work in the DAWN environment).
(savu_v1.2) ~>module add hdfview Loading 64 bit hdfview (savu_v1.2) ~> (savu_v1.2) ~>hdfview /dls/i13/data/2017/cm16786-1/processing/savu/vxu94780/savu_process_lists/tomo_recon_base_config.nxs & [1] 5394 (savu_v1.2) ~>
>>> mod 1.4 ['<dark_file>', '<dark_path>', 1]
and, correspondingly,
>>> mod 1.1 ['<flat_file>', '<flat_path>', 1]
For example, the tomo-centre example given on Reconstruction from image data in the HDF format: the tomo-centre and tomo-recon commands can be reproduced by mod-ing the NxtomoLoader process (<process-index> = 1) for the presence of external darks and flats as follows:
Appendix F: Viewing of Savu output files containing final-reconstruction slices |
|---|
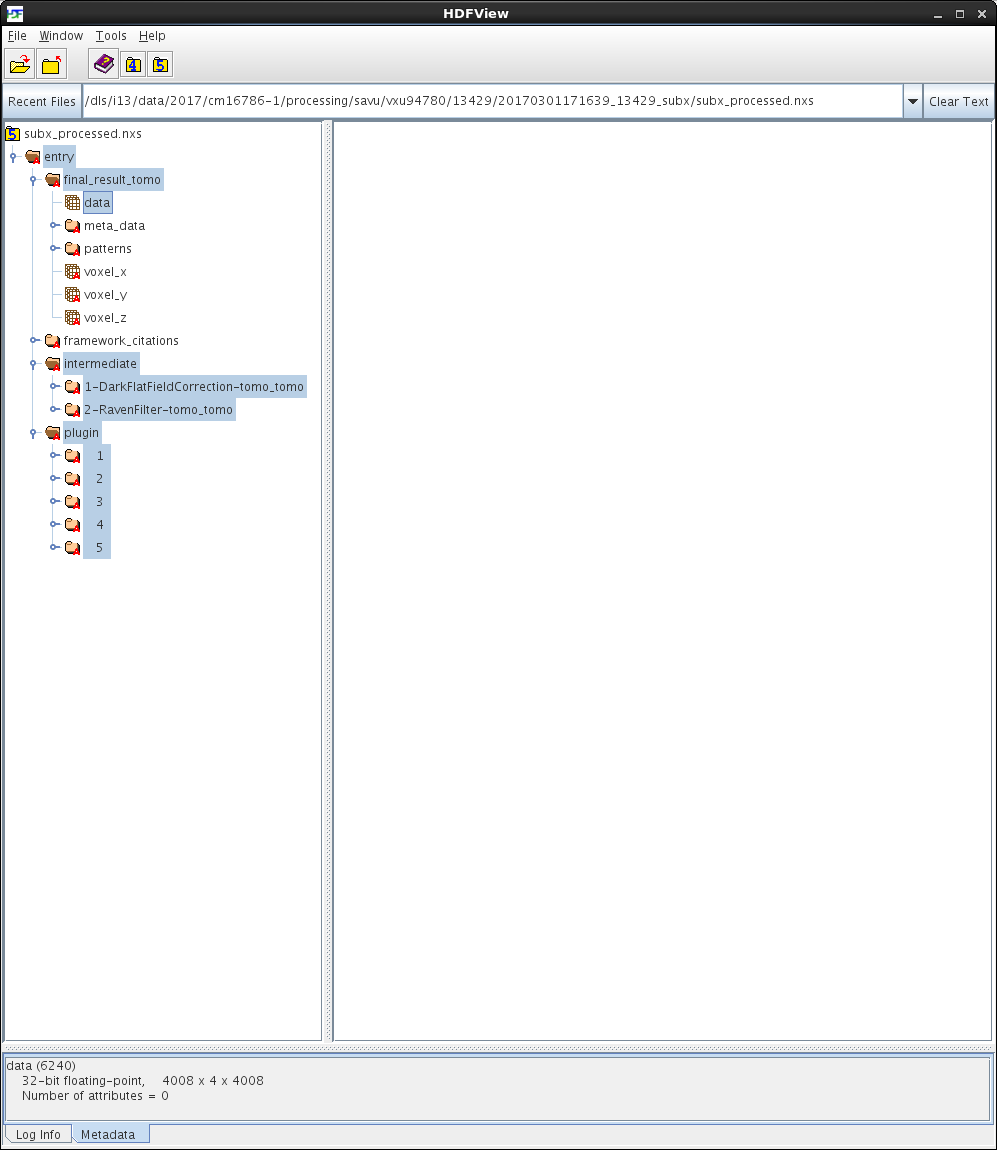
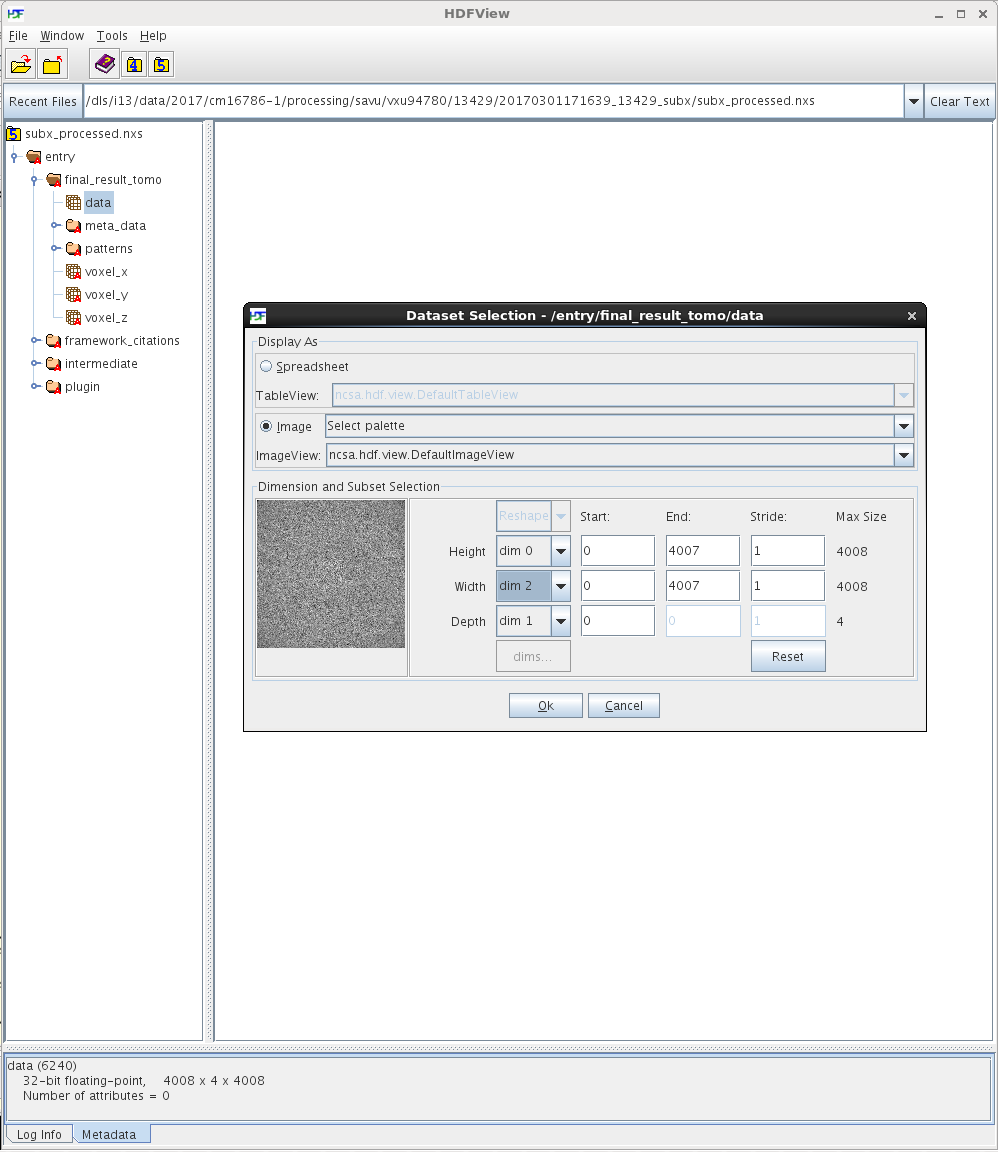
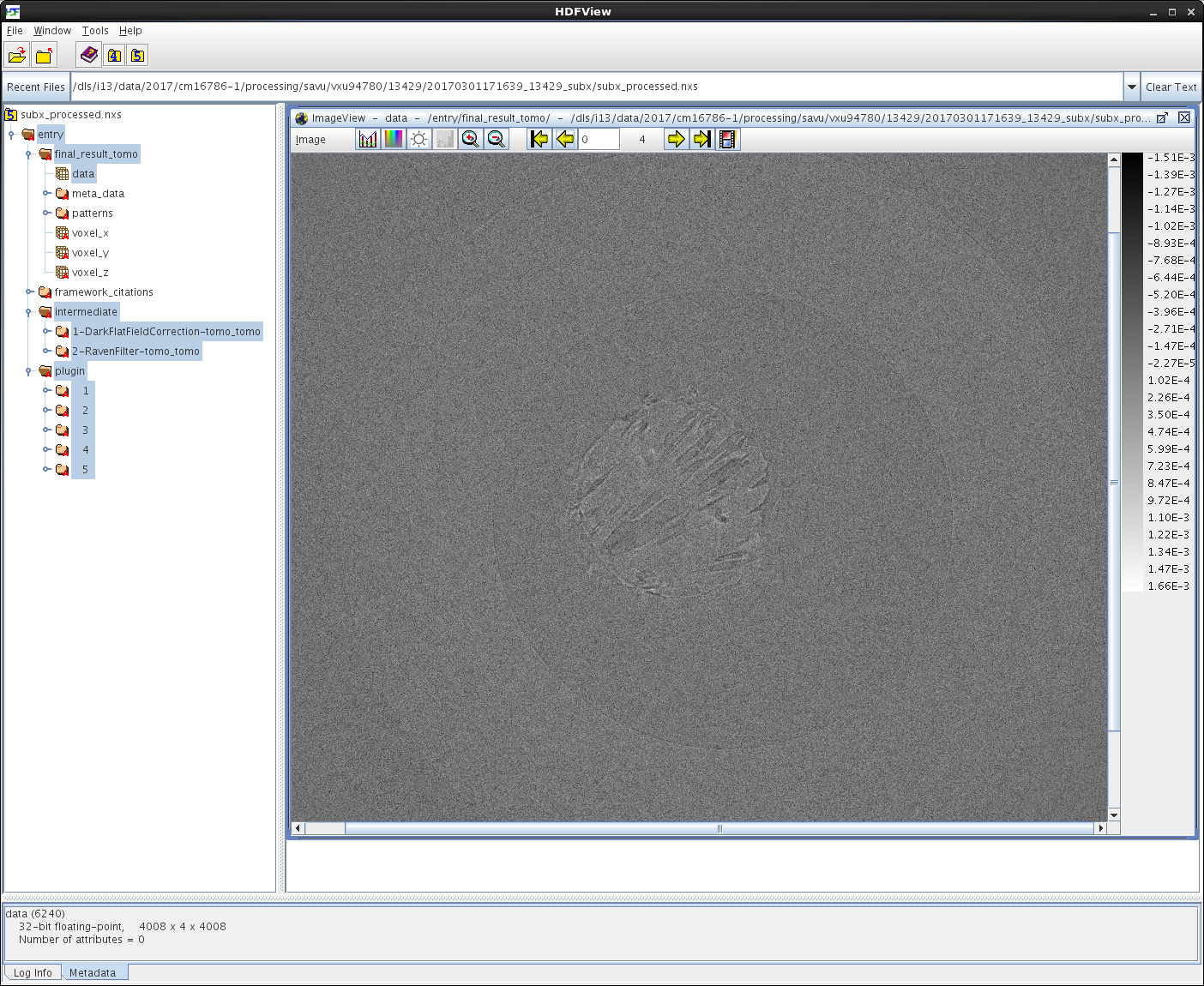
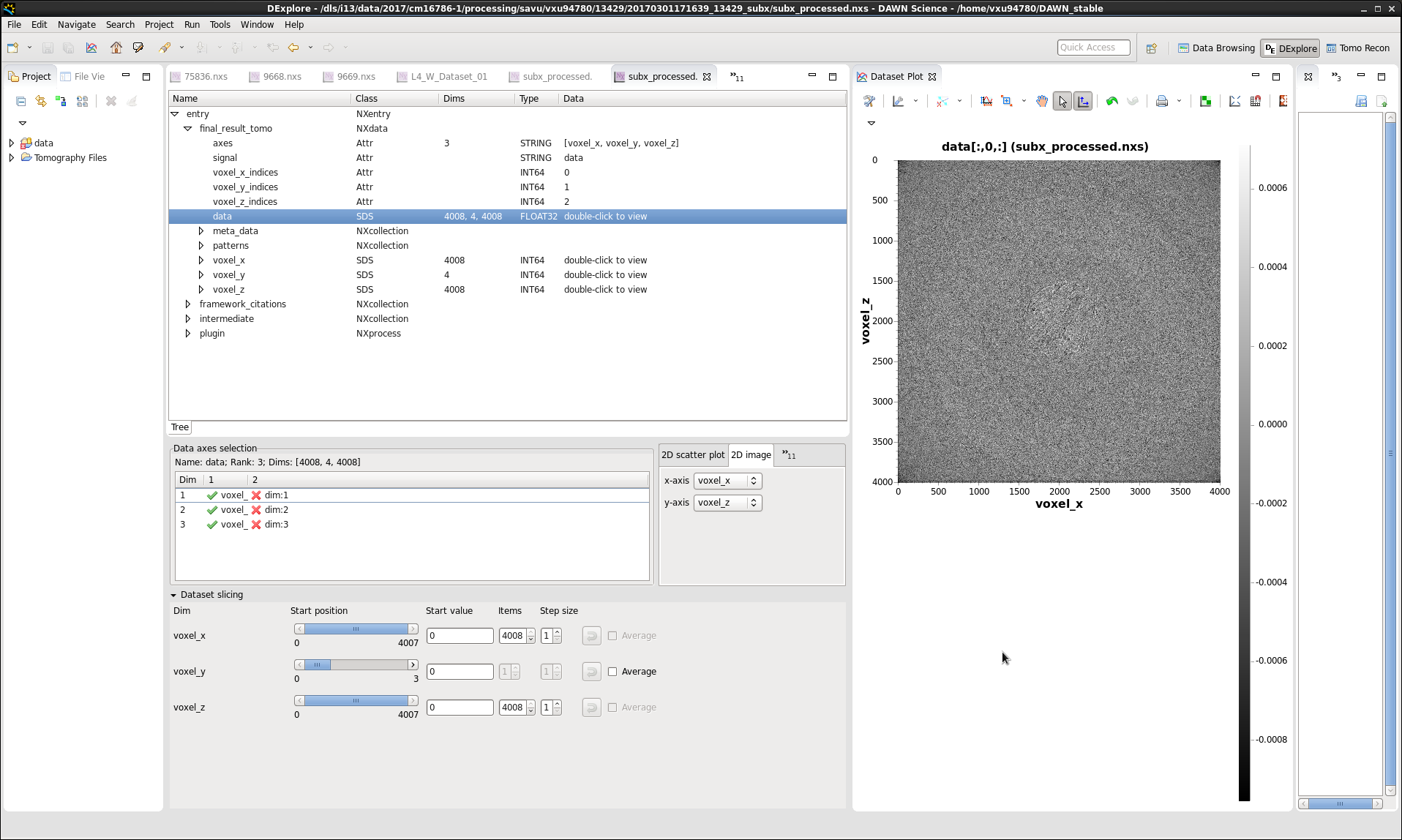
Note that the shape of the /entry/final_result_tomo/data dataset in subx_processed.nxs is 4008 x 4 x 4008, where the number 4 corresponds to the total number of final-reconstruction images, as expected. To view the /entry/final_result_tomo/data dataset as individual images, first left-click on the dataset to select it, then right-click on it and choose the Open As option from the menu, and finally configure this option as illustrated on the screenshot below:
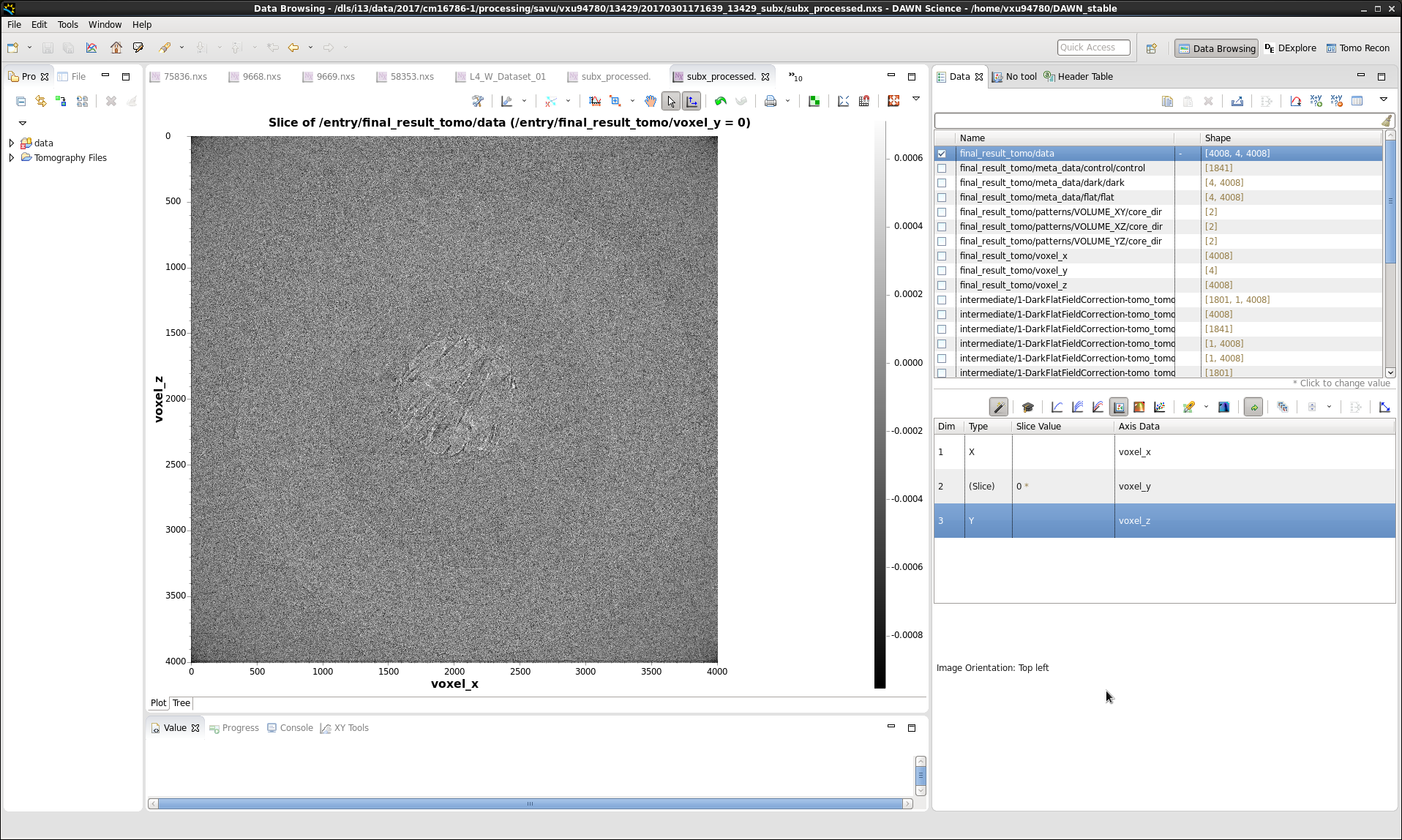
Note that viewing of images stored in HDF5 datasets is most conveniently done in DAWN. For information on how to use DAWN, see DAWN Tutorials. The screenshots of the Data-Browsing and DExplore perspectives are shown below:
Appendix G: Other frequently used Savu-Configurator commands |
|---|
list <plug-in-category>
Use the list command with an appropriate category qualifier to display a list of all plug-ins in this category that are available for use in Savu, i.e. corrections, filters, loaders, reconstructions, or savers.
list <correction[s] | filter[s] | loader[s] | reconstruction[s] | saver[s]>
For example, the following reconstruction options were available in Savu at the time of writing these words (Wed 1 March 2017):
>>> list reconstruction ----------------------------------------- AstraReconCpu Wrapper around the Astra toolbox for cpu reconstruction AstraReconGpu Wrapper around the Astra toolbox for gpu reconstruction CglsRecon Wrapper around the CCPi cgls reconstruction ScikitimageFilterBackProjection Wrapper for scikitimage FBP function ScikitimageSart Wrapper for scikitimage SART function SimpleRecon A simple implementation of a reconstruction routine for testing TomopyRecon Wrapper around the tomopy reconstruction algorithms. See ' http://tomopy.readthedocs.io/en/latest/api/tomopy.recon.al gorithm.html' ----------------------------------------- >>> >>> list corrections ----------------------------------------- DarkFlatFieldCorrection A Plugin to apply a simple dark and flatfield correction to raw timeseries data McNearAbsorptionCorrection A plugin apply hogans xrf absorption correction using stxm data TimeBasedCorrection A time-based dark and flat field correction using linear interpolation TimeBasedPlusDriftCorrection A time-based dark and flat field correction that accounts for image drift (Note: A work in progress but please try) ----------------------------------------- >>>
list <any-characters>*
Use the list command with the trailing asterisk (*) wildcard to filter through any relevant Savu plug-ins. For example, there is no need to memorise the precise (case-sensitive) name of the Raven correction in Savu or the name of the NxtomoLoader loader because their exact, full names can conveniently be identified with the aid of their approximate, 'lazy' names as follows:
>>> list rav* ----------------------------------------- RavenFilter A plugin to remove ring artefacts ----------------------------------------- >>> >>> list nxto* ----------------------------------------- NxtomoLoader A class for loading standard tomography data ----------------------------------------- >>>
disp -q
Use the disp -q command for quiet and quick display of the contents of the currently active provess list in Savu Configurator:
>>> disp -q 1) NxtomoLoader 2) DarkFlatFieldCorrection 3) RavenFilter 4) AstraReconGpu 5) Hdf5TomoSaver >>>
history
Use the history command to list the previously typed-in commands in Savu Configurator, e.g.
1 : help 2 : disp 3 : list nxto* 4 : add NxtomoLoader 5 : mod 1.6 [:, mid-5:mid+5, :] 6 : list dark* 7 : add DarkFlatFieldCorrection 8 : list rav* 9 : add RavenFilter 10 : list pag* 11 : add PaganinFilter 12 : mod 4.off 13 : disp -q 14 : list vo* 15 : add VoCentering 16 : list astra* 17 : add AstraReconGpu 18 : list saver 19 : add Hdf5TomoSaver 20 : disp 3 -v 21 : mod 3.6 50 22 : disp 23 : save tomo_recon_preview.nxs 24 : y
move <from-process-index> <to-process-index>
Use the move <from-process-index> <to-process-index> command to swap the places of any 2 processes in the currently active provess list in Savu Configurator. For example, one can first invoke the add command to attach the VoCentering process (<process index> = 6) to the end of the basic process list of 5 processes, and then move this process to its more appropriate position in the chain, situated immediately before the AstraReconGpu process (<process index> = 4) :
Appendix H: Savu Tips & Tricks |
|---|
- The contents of any Nexus file (generated by Savu) are best viewed in DAWN, using either the File Browsing or DExplore perspective. However, if you intend to use DAWN in the same Linux terminal as Savu, then remember that it is essential to execute 'module add savu' after 'module add dawn' (as otherwise Savu might not work in the DAWN environment).
- Use the savu_mpi command with either the -d or the --tmp option to save any intermediate, auxiliary files to a temporary, periodically-purged directory, e.g. /dls/i13/data/2017/cm16786-1/tmp/ or, more generally, /dls/<ixx>/data/<YYYY>/<visit-id>/tmp/. For more information about this option, execute the savu -h command.
If neither destination path nor filename are provided on invoking the save command on the currently-open process list in Savu Configurator, then this process list gets saved in (overwrites) the original process-list file (i.e. the file whose path was supplied on invoking the preceding open command), e.g.
If filename only (without an absolute or relative path) is provided on invoking the save command on the currently-open process list in Savu Configurator, then this process list gets saved under that filename in the current working directory, e.g.
Apart from the dataset containing reconstructed slices (i.e. /entry/final_result_tomo/data), Savu's principal output file, <Nexus-scan-filename>_processed.nxs, contains all details of the process list that was used to generate these reconstructions. This can be quite convenient, for example, to retrieve or verify the value of CoR or any other reconstruction parameter that was used in a particular reconstruction chain. In addition, one can invoke the open command directly on the <Nexus-scan-filename>_processed.nxs file to load its internally-stored process list into Savu Configurator, and then use this process list, or perhaps its modified version, to reconstruct another tomography dataset, stored in a different Nexus scan file.
- cntd.